This year has shaped up to be a busy but exciting year, and one of these exciting parts has been the opportunity to publish some of my work in the human microbiome/virome space. A couple of weeks ago our research group (from when I was at the University of Michigan) had our colon cancer virome preprint fully published in mBio, and I am really excited to see it out. For this post I want to highlight that work and provide a little extra commentary about how I think it fits in the field and where the work is going.
Our recent paper was titled “Diagnostic Potential and Interactive Dynamics of the Colorectal Cancer Virome” and is freely available here on the mBio website. Briefly, we resequenced a frozen archived stool sample set from the previously published paper from the same lab. The original Zackular study utilized 16S rRNA shotgun sequencing, but we went back and performed shotgun metagenomic sequencing on both the whole stool as well as VLP (virus-like particle) purified stool. We found that altered viral communities were associated with the cancer state, and that this signal was dominated by bacteriophages (bacterial specific viruses). The bacteriophages were not associated with particular pathogens such as Fusobacterium nucleatum. This observation was interesting, but how do we interpret an association between bacteriophages and colon cancer?
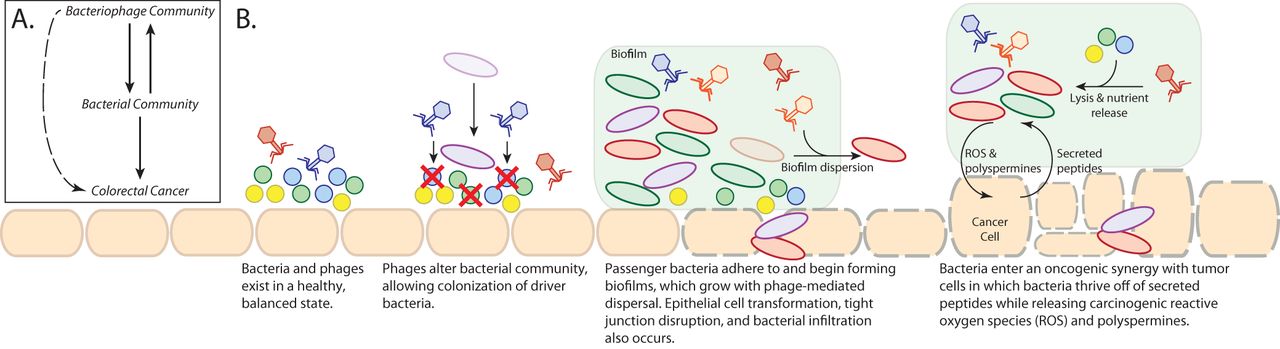
There are different ways in which the bacteriophage communities could impact colorectal cancer progression, and we outlined some of those in a working hypothesis diagram (see figure above). Based on our findings and previous publications, we hypothesize the phages may be indirectly impacting colorectal cancer progression by contributing to the bacterial community dynamics. The bacteriophages could be causing the initial shifts in bacterial community composition, which in turn contribute to initial cancer progression. The phages could also be contributing to the cancer-associated biofilm dynamics and structure, as well as bacterial lysis and nutrient availability. It is interesting to consider these results in the context of previous literature that outlines the impact of different antibiotics on colorectal cancer progression. Because phages can also kill bacteria, they may impact cancer progression through a similar mechanism as what was observed in the different antibiotic treatments. There are many exciting and important directions in which this work can be taken.
At approximately the same time we published this work out of preprint, the group of Nakatsu et al published another study which also looked at the gut virome association with colorectal cancer (paper can be found here). This work offers some interesting additional perspectives to the underlying biology of our system. A big caveat to comparing the two studies is that the group used whole shotgun metagenomic sequencing (no VLP purification) and targeted their bioinformatics analysis toward virus reference genomes. This was unlike our Hannigan et al paper that performed VLP purification and more of a de novo analysis approach. Despite the differences between the studies, both identified significant gut virome signatures associated with colorectal cancer, and together they provide compelling evidence for a role of the gut virome in colorectal cancer.
Not only am I excited about the work our group recently published, I am also very excited about the work coming out of other research groups and the overall direction of the field. In addition to these types of sequencing studies, I expect to see some follow up “mechanistic” work to address hypotheses like those we outlined above. Like other areas of microbiome research, I also expect to see future work begin to study signatures associated with microbes such as fungi or maybe even archaea. This will continue to be an interesting and medically important field to watch.
comments powered by Disqus