Understanding and elucidating the links between the microbiome and human health is a complex problem, largely due to the complicated webs of interactions between members of the microbiome and the human host. One layer of this complicated system is the functionality of bacteria within the microbiome. Small molecules, often produced by microbial biosynthetic gene clusters (BGCs), are an avenue many groups are now taking to further elucidate this functionality. As we discussed in this blog before, BGCs are clusters of co-localized genes that together encode the “machinery” needed to produce small molecules that can impact functionality of either human cells or other microbial cells. In this blog post we will discuss the recently reported work of Aleti et al, in which they reported an analysis of the biosynthetic potential of the oral microbiome, and its association with oral disease. The group’s disease focus was on periodontitis and dental plaques, which are significant disease burdens.
To study the biosynthetic gene clusters in the oral microbiome, the researchers generated a database of 461 oral bacteria reference genomes and used the AntiSMASH infrastructure to identify BGCs within those genomes. The majority of BGC classes were unsurprisingly unclassifiable, although the group was able to infer classes of some of those BGCs by aligning them to the BGCs within the MiBIG database of 1400 reference BGCs.
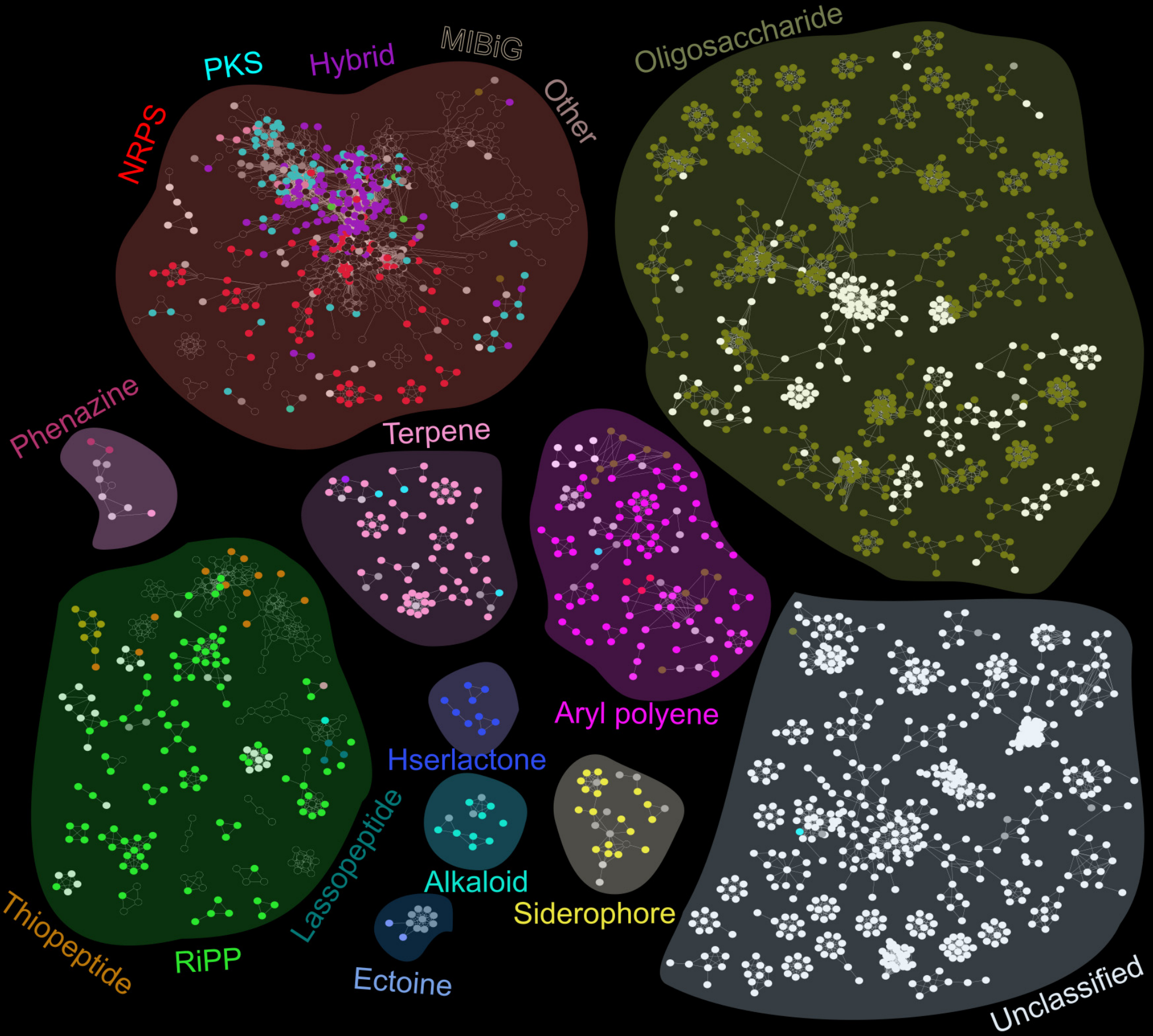
Once their BGC reference set was complete, the scientists curated a dataset of metagenomic (DNA; functional potential) and meta-transcriptomic (RNA; functional gene expression) information from saliva and dental plaques of 294 subjects who were either healthy, had dental caries, or periodontitis. Alignment of the metagenomic and meta-transcriptomic sequences to the BGC database allowed the group to identify differentially abundant/expressed BGCs between the disease states. The group was also able to follow up with some metabolic analysis (using LC-MS/MS) to find and validate some signals in the microbiome. Finally, a co-abundance network analysis revealed numerous correlations between BGCs and bacterial taxonomic groups. Most correlations were negative, which the authors suggest reflects antagonistic antimicrobial BGC relationships.
This is an interesting study that highlights the prevalence of BGCs within the oral microbiome, and the association of those BGCs with oral disease states. The specific disease-associated BGC classes will likely inform hypotheses for the field (these details are beyond the scope of this blog post and can be read in the original manuscript). This paper is particularly notable for how it sets up future studies. In addition to predicting BGCs from reference genomes, it would valuable for future studies to apply the supplemental approach of predicting BGCs from de novo assemblies from the metagenomic data. Additionally, it would be good to focus on the BGCs that are associated with other interactions beyond those with bacterial taxa. BGC associations with host functions and other microbes, such as fungi, would offer a new level of understanding to the complex system.
This manuscript was published in the journal mBio and is therefore open to be read by anybody (find it here). It is a quick read and could be a useful reference if you are in this field, so go ahead, download it, and check it out. As always, if you have any questions, comments, or concerns, please don’t hesitate to drop me a note in the comment section. Thanks for reading!
comments powered by Disqus