Introduction
We make a lot of logical connections as we do science. In biology we discover new relationships, new biological pathways, and new ways that organisms interact. Given the accelerated rise in available scientific data and the resulting connections between that data, we may ask a simple question: how can we make sense of it all? Connecting the dots across our vast understanding of biology is no small task, and researchers have increasingly turned to knowledge graphs as a solution.
What Is A Knowledge Graph?
Having seen growing interest and attention in recent years, knowledge graphs are a popular approach to gleaning valuable insights from complex data, especially in biological research. In the biological sciences, researchers are leveraging knowledge graphs to understand mechanisms, discover new medicines, and solving other valuable problems. However before going further, it’s important for us to first understand what a knowledge graph is.
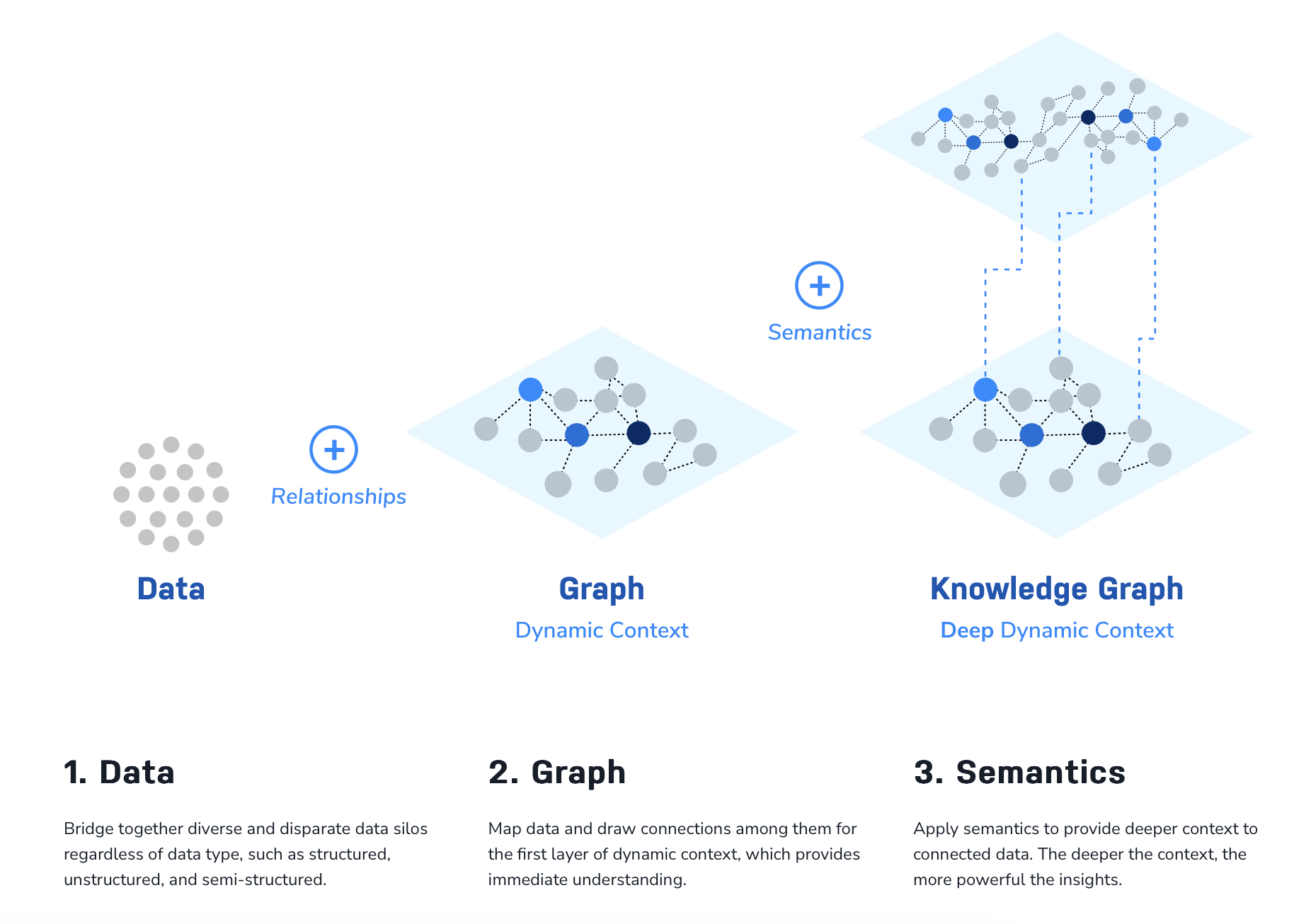
To define a knowledge graph, we can turn to (arguably) one of the leading graph technology platforms: Neo4j. Neo4j defines a knowledge graph as “an interconnected dataset enriched with semantics so we can reason about the underlying data and use it confidently for complex decision-making” (check out the source for a full description). As outlined in the Neo4j figure below, a graph database allows us to connect data with relationships while a knowledge graph overlays meaning onto the graph data (through semantics). This layer of knowledge allows us to glean deeper insights into our data and can better inform decision-making and scientific insights.
Using Knowledge Graphs In Biology
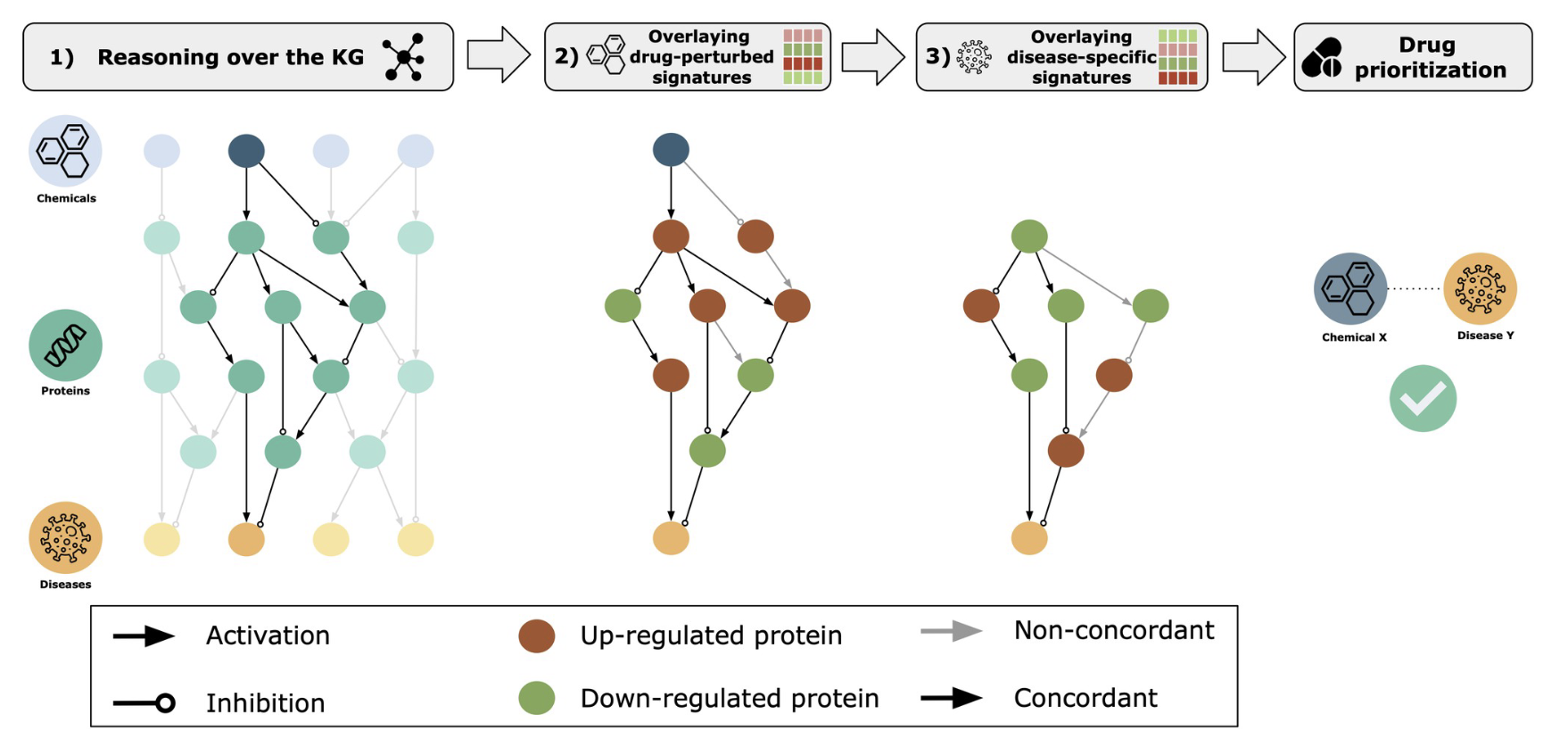
The knowledge graph field is rich with information and value continues to be shown across diverse industries including finance, manufacturing, and biotech. A fascinating and promising area for applying knowledge graphs is in drug discovery and development. One research article that articulates this well is “Causal reasoning over knowledge graphs leveraging drug-perturbed and disease-specific transcriptomic signatures for drug discovery” by Domingo-Fernandez, et al (2022). In this article, the authors outline their method RPath which they describe as “a novel algorithm that prioritizes drugs for a given disease by reasoning over causal paths in a knowledge graph”. Essentially this method aims to empower researchers to glean new insights in biology and therapeutics.
The authors of this paper begin by building out a knowledge graph which layers causal relationships (activator and inhibitory) on top of a graph of chemical, protein, and disease data. As outlined in their figure below, the authors then build their algorithm to link chemicals to diseases as a way to prioritize drug discovery and validation. The authors incorporate transcriptomic signatures into this as well, and we can read the paper for those interesting details.
One value of this approach is also its scalability. We could imagine many ways in which new data layers could be added to the database, and then how the resulting knowledge could be leveraged. We can also think about many more sophisticated ways to analyze the data contained in the knowledge graph, such as graph neural networks or other machine learning approaches.
Conclusion
Knowledge graphs have been seeing an astounding growth in popularity and interest, and for good reason. Connecting large and complex data graphs with contextual and semantic relationships allows us to glean deeper insights into data that can drive more value across industries. In the biological sciences, we can use knowledge graphs to better understand disease and use that information to inform therapeutic discovery and development. This field continues to show great promise and has already demonstrated exciting results. It will be exciting to watch this field evolve.
comments powered by Disqus